Artificial Evolution
Alongside studying the evolution of duplicated genomes through experimental evolution, we also investigate the evolution of polyploid genomes using computer-based in-silico simulations. These simulations involve tracking the evolution of (duplicated) artificial genomes that encode gene regulatory networks, providing valuable insights into the dynamics and adaptation of polyploid systems.
In our lab, we have build our own artificial framework to mimic biological evolution. In this framework, every Digital Organism (DO) has its own genome, which has been inspired by the model previously proposed by Reil (1999) and consists of a randomly created string of digits. Genes are not pre-specified, but identified in the randomly built genome. Compared to the initial model of Reil, our model makes an explicit distinction between regulatory and structural genes. Upon initialization of a new DO, specific genes will be activated, dependent on certain cues of the environment. After activation or ‘expression’ of both regulatory and structural genes, these will ‘produce’ so-called embedded ‘agents’ that together will form a Gene Regulatory Network (GRN) and exert a certain control of the DO. Each of the DOs has the same virtual functionalities and sensors to allow them to interact with each other and to respond to the environment. In our simulations, energy sources (representing food for the DOs) are distributed over a grid. These food sources can ‘grow’ their energy level gradually and replicate more food with a certain rate. At every time step, DOs consume energy for living and need to constantly retrieve energy from the environment to survive. Evaluation of fitness and adaptation of the DOs swarm/population as a whole is then based on their overall energy level. The implementation of necessary functions for the DO to survive and replicate requires a certain amount of energy and high energy levels help DOs compete with others. However, higher energy levels do not always equal better adaptability of the DO. True adaptability of the DOs is dependent on both the interaction with the environment and other DOs. For example, consuming too much food at one time will lead to less food left to grow more food at later stages. Although such behavior will increase the energy of the DOs on the short term, it will lead to extinction or harsh selection later. Using populations of DOs should give us the freedom to explore a wide plethora of options to investigate when and under which circumstances polyploidy or WGD can pose a selective advantage to the organism or population. For instance, based on the large amount of information that has been gained already from comparative genomics approaches using real genomes that have undergone genome-wide duplications, we can model and/or assess the effect of duplications on the structure, evolution and rewiring of GRNs.
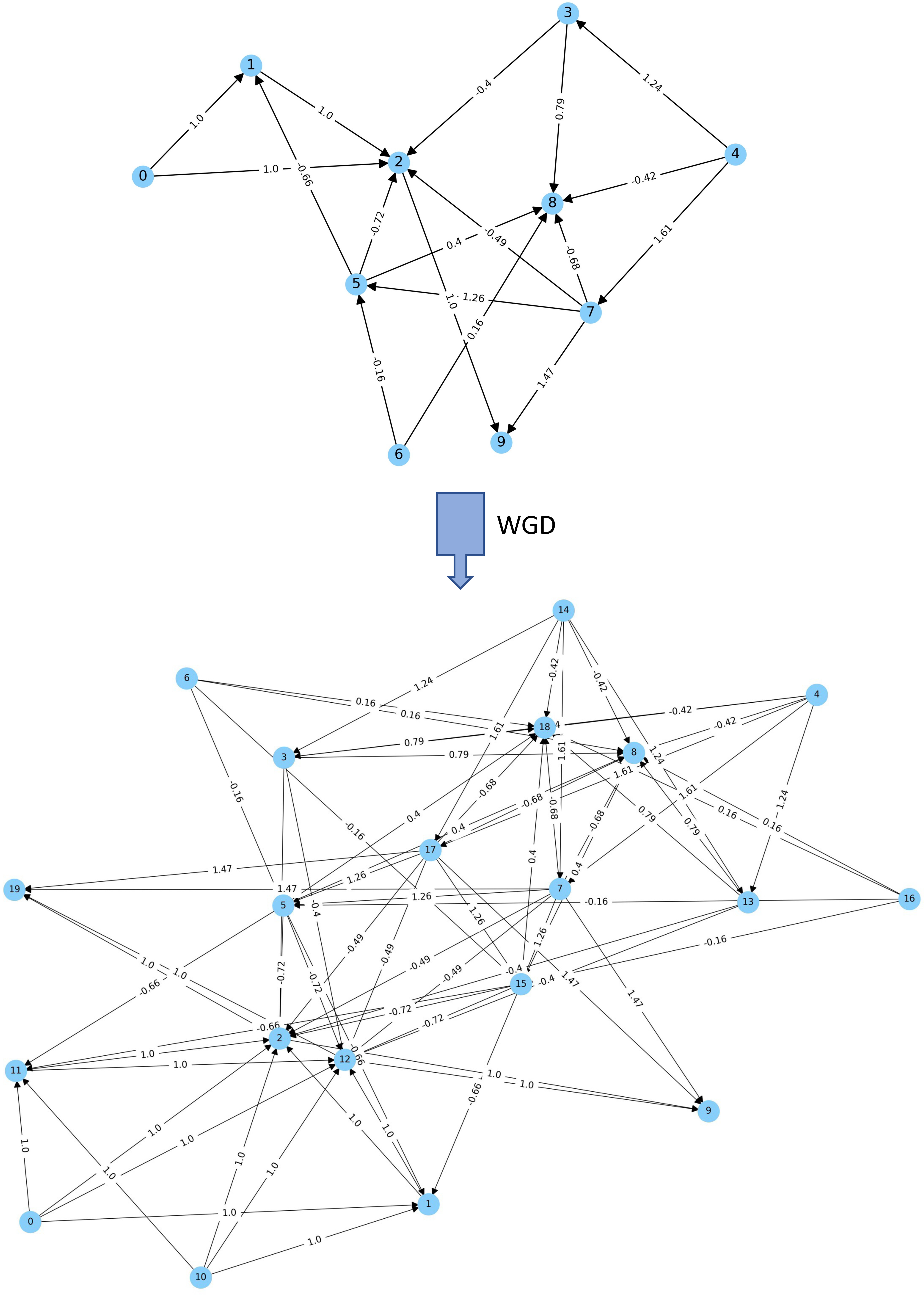
Being able to replay the ‘duplication-tape-of-life’ through hundreds or thousands of simulations, and changing different parameters such as the birth and death rates of genes, biased retention of genes subsequent to duplication events, dosage effects, etc., we will be able to study the (at least theoretical) effects of WGDs on mutational and environmental robustness.
Our initial work on bio-inspired agent-based modelling has demonstrated the usefulness of our computational simulation in studying the role of WGD in evolution and adaptation, helping to overcome some of the traditional limitations of evolution experiments with model organisms. For example, we found that, under stable environments, DOs with non-duplicated genomes formed the majority, if not all, of the population, whereas the numbers of DOs with duplicated genomes increased under dramatically challenging environments. After tracking the evolutionary trajectories of individual genomes in terms of sequence and encoded gene regulatory networks (GRNs), we propose that duplicated GRNs might provide polyploids with better chances to acquire the drastic changes necessary to adapt to challenging conditions, thus endowing DOs with increased adaptive potential under extinction events. In contrast, under stable environments, random mutations might easily render the GRN less well adapted to such environments, a phenomenon that is exacerbated in duplicated, more complex GRNs.
More recently, we expanded this approach by a deeper investigation of duplicated artificially generated Gene Regulatory Networks (aGRNs). We showed that such duplicated aGRNs—and thus duplicated genomes—show higher signal output variation than non-duplicated aGRNs. This increased variation leads to niche expansion and can provide polyploid populations with substantial advantages to survive environmental turmoil. In contrast, under stable environments, GRNs might be maladaptive to changes, a phenomenon that is exacerbated in duplicated GRNs. We believe that these results provide insights into how genome duplication and (auto)polyploidy might help organisms to adapt quickly to novel conditions and to survive ecological uproar or even cataclysmic events.
Published papers:
- Yao, Y., Marchal, K., & Van de Peer, Y. (2014). Improving the adaptability of simulated evolutionary swarm robots in dynamically changing environments. PLoS One, 9(3), e90695.
- Yao, Y., Storme, V., Marchal, K., & Van de Peer, Y. (2016). Emergent adaptive behaviour of GRN-controlled simulated robots in a changing environment. PeerJ, 4, e2812.
- Yao, Y., Carretero-Paulet, L., & Van de Peer, Y. (2019). Using digital organisms to study the evolutionary consequences of whole genome duplication and polyploidy. PloS one, 14(7), e0220257.
- Ebadi, M., Bafort, Q., Mizrachi, E., Audenaert, P., Simoens, P., Van Montagu, M., ... & Van de Peer, Y. (2023). The duplication of genomes and genetic networks and its potential for evolutionary adaptation and survival during environmental turmoil. Proceedings of the National Academy of Sciences, 120(41), e2307289120.