Experimental Evolution
At present, knowledge about the (eco-)evolutionary effects of polyploidy is largely based on either the comparison of contemporary polyploids with their diploid ancestors' descendants, or based on signatures of Whole Genome Duplications (WGDs) in present-day plant genomes. These comparative methods, although informative, suffer from potential biases introduced by evolution of both the polyploid and the descendants of its ancestors after establishment. To gain deeper mechanistic insights into the short and long-term effects of polyploidy, experimental studies comparing synthetic polyploids and their direct, lower-ploidy ancestors are essential. We have initiated several (eco-)evolutionary experiments wherein both conditions for initial establishment and long(er)-term evolution of polyploid genome characteristics can be studied.
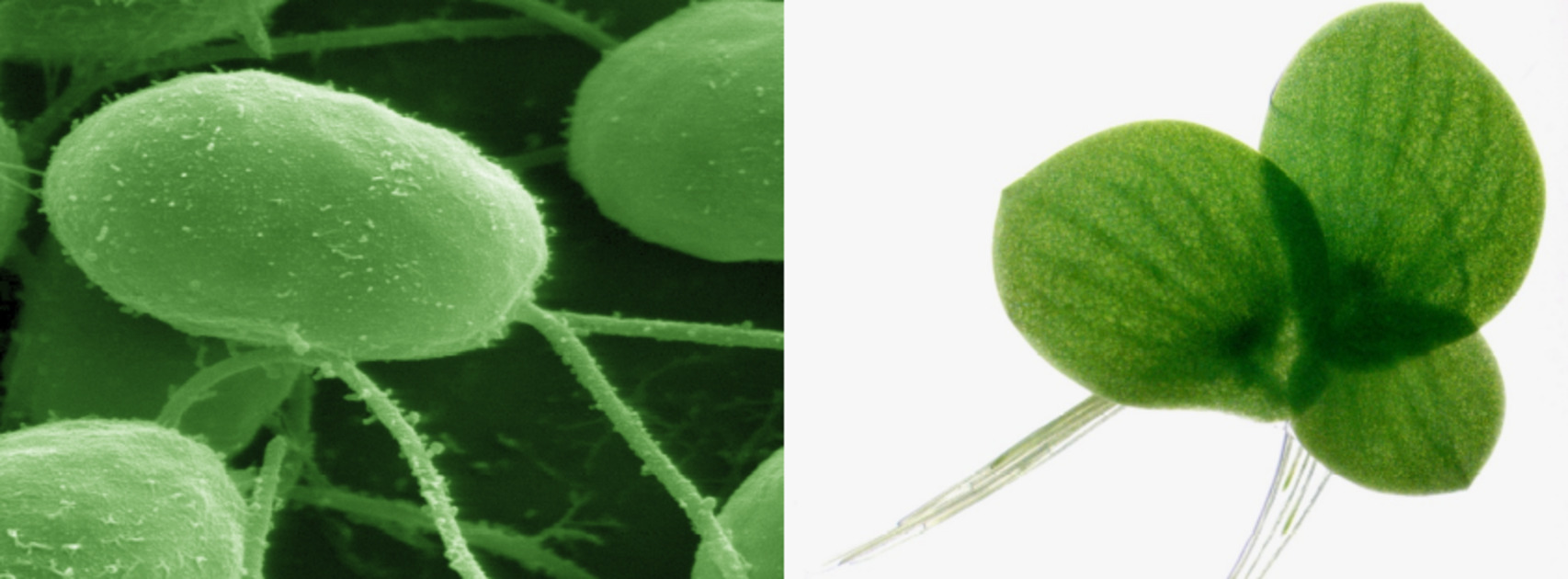
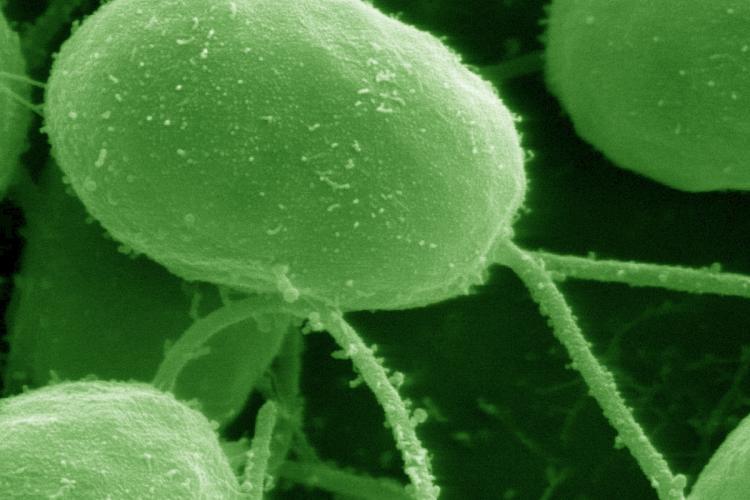
Because polyploidy seems to be especially preponderant in the green lineage, we use the unicellular green alga Chlamydomonas reinhardtii and the small aquatic macrophyte Spirodela polyrhiza to study polyploidization in the green lineage. Both organisms are characterized by a high vegetative growth rate, relatively small genome sizes, limited space requirements, easy culture establishment in the laboratory, and a rich history as a model system, making them well suited to study the functional effects of WGD and long term evolution.
Chlamydomonas reinhardtii
Chlamydomonas reinhardtii, a haploid green alga, is a valuable model organism with profound potential to study the impact of ploidy increase on the longer-term in a laboratory environment.
You can find out more about how to use Chlamydomonas to study polyploidy in our book chapter and one of our papers.
We use Chlamydomonas to study:
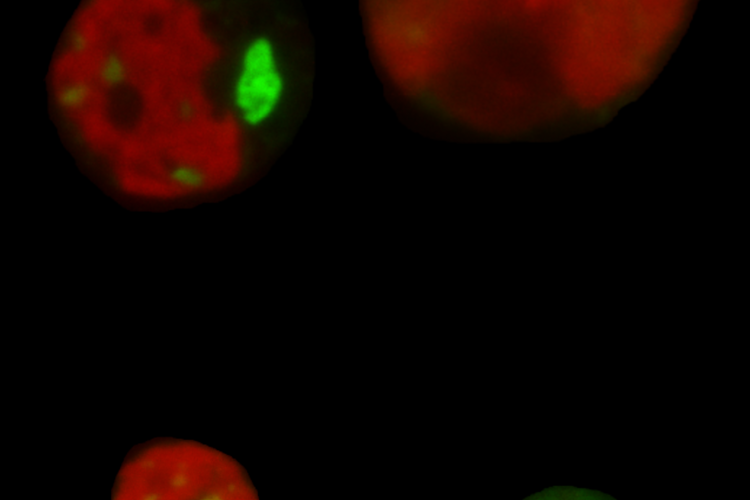
- The transcriptomic response to allopolyploidy. See:
- Differences in adaptation rate between diploid and haploid lines
- Ploidy induced changes in mutation rate and spectrum using mutation accumulation
- Competition between cells of different ploidy levels
- Genome downsizing after WGD
Spirodela polyrhiza

Alongside Chlamydomonas reinhardtii, we use the small aquatic flowering plant Spirodela polyrhiza (greater duckweed) for our evolutionary experiments. Because polyploidy is most commonly found in flowering plants, and the ancestors of duckweeds have experienced two whole-genome duplications (WGDs) (after which they rediploidized), it serves as an excellent model system for studying polyploidy.
You can learn more about duckweeds on our (co-)created website: thecharmofduckweed.org. Moreover, you can find out how to use Spirodela to study polyploidy in our book chapter.
In our duckweed research, we focus on several aspects of polyploid evolution:
- The immediate phenotypic and molecular effects of WGD in genetically diverse Spirodela clones. See:
- Bafort, Q., Wu, T., Natran, A., De Clerck, O., & Van de Peer, Y. (2023). The immediate effects of polyploidization of Spirodela polyrhiza change in a strain-specific way along environmental gradients. Evolution Letters, 7(1), 37-47.
- Wu, T., Bafort, Q., Mortier, F., Almeida‐Silva, F., Natran, A., & Van de Peer, Y. (2024). The immediate metabolomic effects of whole‐genome duplication in the greater duckweed, Spirodela polyrhiza. American Journal of Botany, 111(8), e16383.
- Population dynamics and establishment of tetraploids in a diploid population by having tetraploids compete directly with their diploid progenitor populations. See:
- Ploidy induced changes in mutation rate and spectrum using mutation accumulation
- Molecular and rate differences in the adaptation to a stressful environment using longer-term laboratory evolution experiments
- Differences in metabolic rate between tetraploids and diploids
- Natural occurrence of polyploidy in the duckweed family